Sensitivity Analysis¶
GloMPO implements a sensitivity analysis method to aid in reducing the dimensionality of high-dimensional optimization problems. The Hilbert-Schmidt Independence Criterion (HSIC) is the chosen method due to its robustness and accuracy.
The analysis module comes with three main components:
Class |
Description |
|---|---|
Performs the sensitivity calculation |
|
Result of a sensitivity calculation |
|
Base class of the kernels needed by |
HSIC¶
-
class
HSIC(x, y, labels=None, x_bounds='auto')[source]¶ Implementation of the Hilbert-Schmidt Independence Criterion for global sensitivity analysis. Estimates the HSIC via the unbiased estimator of Song et al (2012).
- Parameters
- x
\(n \times d\) matrix of \(d\)-dimensional vectors in the input space. These are samples of the variables you would like to know the sensitivity for.
- y
\(n\) length vector of outputs corresponding to the input samples in
x. These are the responses of some function against which the sensitivity is measured.- labels
Optional list of factor names.
- x_bounds
\(d \times 2\) matrix of min-max pairs for every factor in
xto scale the values between 0 and 1. Defaults to'auto'which takes the limits as the min and max values of the sample data. To use no scaling set this toNone.
- Notes
yis automatically scaled by taking the logarithm of the values and then scaling by the minimum and maximum to a range between zero and one. This makes selecting kernel parameters easier, and deals with order-of-magnitude problems which often arise during reparameterization.- References
Song, L., Smola, A., Gretton, A., Bedo, J., & Borgwardt, K. (2012). Feature Selection via Dependence Maximization. Journal of Machine Learning Research, 13, 1393–1434. https://doi.org/10.5555/2188385.2343691
Gretton, A., Bousquet, O., Smola, A., & Schölkopf, B. (2005). Measuring Statistical Dependence with Hilbert-Schmidt Norms. In Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics): Vol. 3734 LNAI (Issue 140, pp. 63–77). Springer, Berlin, Heidelberg. https://doi.org/10.1007/11564089_7
Gretton, A., Borgwardt, K. M., Rasch, M. J., Smola, A., Schölkopf, B., Smola GRETTON, A., & Smola, A. (2012). A kernel two-sample test. Journal of Machine Learning Research, 13(25), 723–773. http://jmlr.org/papers/v13/gretton12a.html
Spagnol, A., Riche, R. Le, & Veiga, S. Da. (2019). Global Sensitivity Analysis for Optimization with Variable Selection. SIAM/ASA Journal on Uncertainty Quantification, 7(2), 417–443. https://doi.org/10.1137/18M1167978
Da Veiga, S. (2015). Global sensitivity analysis with dependence measures. Journal of Statistical Computation and Simulation, 85(7), 1283–1305. https://doi.org/10.1080/00949655.2014.945932
-
compute(inputs_kernel=None, outputs_kernel=None, n_bootstraps=1, n_sample=- 1, replace=False)[source]¶ Calculates the HSIC for each input factor. The larger the HSIC value for a parameter, the more sensitive
yis to changes in the corresponding parameter inx.- Parameters
- inputs_kernel
Instance of
BaseKernelwhich will be applied tox. Defaults toGaussianKernel.- outputs_kernel
Instance of
BaseKernelwhich will be applied toy. Defaults toConjunctiveGaussianKernel.- n_bootstraps
Number of repeats of the calculation with different sub-samples from the data set. A small spread from a large number of bootstraps provides confidence on the estimation of the sensitivity.
- n_sample
Number of vectors in
xto use in the calculation. Defaults to -1 which uses all available points.- replace
If
True, samples fromxwill be done with replacement and vice verse. This only has an effect ifn_sampleis less thannotherwise replace isTrueby necessity.
- Returns
HSICResultObject containing the results of the HSIC calculation and the calculation settings.
-
compute_with_reweight(residuals, targets, error_weights, error_sigma, inputs_kernel=None, outputs_kernel=None, n_bootstraps=1, n_sample=- 1, replace=False, max_cache_size=None)[source]¶ Computes the sensitivity of the HSIC to the weights applied to the construction of an error function. This calculation is only applicable in very particular conditions:
\(X \in \mathbb{R}^{n \times d}\) represents the inputs to a function which produces \(m\) outputs for each \(d\) length input vector input of \(X\) resampled \(n\) times.
These outputs are the \(n \times m\)
predictionsmatrix (\(P\)).The
predictionsmatrix can be condensed to the \(n\) length vectoryby an ‘error’ function:\[\mathbf{y} = \sum^d_i w_i \left(\frac{P_i - r_i}{\sigma_i}\right)^2\]\(\mathbf{r}\), \(\mathbf{w}\) and \(\mathbf{\sigma}\) are \(m\) length vectors of
reference,error_weights, anderror_sigmavalues respectively.This function returns \(\frac{dg}{d\mathbf{w}}\) which is a measure of how much changes in \(\mathbf{w}\) (the ‘weights’ in the error function) will affect \(g\) which is a measure of how close HSIC values are to our
targetHSIC sensitivities.\(g\) is defined such that \(g \geq 0\). \(g = 0\) implies the sensitivities are perfect.
- Parameters
- residuals
\(n \times m\) matrix of error values between predictions and references.
- targets
\(d\) length boolean vector. If an element is
True, one would like the corresponding parameter to show sensitivity. Can also send a vector of real values such that sum of elements is 1. This allows for custom sensitivities to be targeted for each parameter.- error_weights
\(m\) length vector of error function ‘weights’ for which the sensitivity will be measured.
- error_sigma
\(m\) length vector of error function ‘standard error’ values.
- inputs_kernel
See
compute(). But only one value is allowed in this function.- outputs_kernel
See
compute().- n_bootstraps
Number of repeats of the calculation with different sub-samples from the data set. A small spread from a large number of bootstraps provides confidence on the estimation of the sensitivity.
- n_sample
Number of vectors in
xto use in the calculation. Defaults to -1 which uses all available points.- replace
If
True, samples fromxwill be done with replacement and vice verse. This only has an effect ifn_sampleis less thannotherwise replace isTrueby necessity.- max_cache_size
Maximum amount of disk space (in bytes) the program may use to store matrices and speed-up the calculation. Defaults to the maximum size of the temporary directory on the system.
- Returns
HSICResultWithReweightObject containing the results of the HSIC calculation and the calculation settings.
Warning
Efforts have been made to reduce the memory footprint of the calculation, but it can become very large, very quickly.
This calculation is also significantly slower than the normal sensitivity calculation.
HSIC Results¶
-
class
HSICResult(*args, **kwargs)[source]¶ Result object of a single HSIC calculation produced by
HSIC.compute(). Can only be created byHSIC.compute()orload().-
property
hsic¶ \(d\) vector of raw HSIC calculation results. If
n_bootstrapsis larger than one, this represents the mean over the bootstraps.- See Also
-
property
hsic_std¶ \(d\) vector of the standard deviation of raw HSIC calculation results across all the bootstraps.
- See Also
-
property
inputs_kernel¶ Name of the kernel applied to the input-space data.
-
property
inputs_kernel_parameters¶ Parameters for the input kernel.
-
classmethod
load(path)[source]¶ Load a calculation result from file.
- Parameters
- path
Path to saved result file
- See Also
-
property
n_bootstraps¶ Number of times the HSIC calculation was performed with different sub-samples of the data.
-
property
n_factors¶ Number of factors analyzed.
-
property
n_samples¶ Number of items in the sub-samples of the data used in each bootstrap.
-
property
order_factors¶ Returns factor indices in descending order of their influence on the outputs. The positions in the array are the rankings, the contents of the array are the factor indices. This is the inverse of
ranking().- Returns
- numpy.ndarray
\(d\) vector of order factors.
- See Also
-
property
outputs_kernel¶ Name of the kernel applied to the output-space data.
-
property
outputs_kernel_parameters¶ Parameters for the output kernel.
-
plot_grouped_sensitivities(path='trend.png', squash_threshold=0.0, _seed_ax=None)[source]¶ Create a pie chart of the
sensitivitiesresult per factor group. Assumeslabelsfor the parameters take the format: group:factor_name.- Parameters
- path
Optional file location in which to save the image. If not provided the image is not saved and only returned.
- squash_threshold
If a group’s sensitivity falls below this value it will be added to the ‘Other’ wedge of the plot.
- Returns
- matplotlib.figure.Figure
Figure instance allowing the user to further tweak and change the plot as desired.
-
plot_sensitivities(path='hsicresult.png', plot_top_n=None)[source]¶ Create a detailed graphic of the
sensitivitiesresults.- Parameters
- path
Optional file location in which to save the image. If not provided the image is not saved and only returned.
- plot_top_n
The number of factors to include in the plot. Only the
plot_top_nmost influential factors are included.
- Returns
- matplotlib.figure.Figure
Figure instance allowing the user to further tweak and change the plot as desired.
-
plot_sensitivity_trends(path='trend.png', _seed_ax=None)[source]¶ Create a summary graphic of the
sensitivitiesresults. This is a more abstract version ofplot_sensitivities()that always includes all parameters. It is useful to inspect the sensitivity imbalance between parameters as well as the spread between repeats.- Parameters
- path
Optional file location in which to save the image. If not provided the image is not saved and only returned.
- Returns
- matplotlib.figure.Figure
Figure instance allowing the user to further tweak and change the plot as desired.
-
property
ranking¶ Returns the ranking of each factor being analyzed. The positions in the array are the factor indices, the contents of the array are rankings such that 1 is the most influential factor and \(g+1\) is the least influential. This is the inverse of
order_factors().- Returns
- numpy.ndarray
\(d\) vector of rankings.
- See Also
-
property
sampling_with_replacement¶ Whether sampling was done with or without replacement from the available data. False, if all available data was used and automatically True if more samples were used than the number available in the data set.
-
save(path='hsiccalc.npz')[source]¶ Saves the result to file. Uses the numpy
'.npz'format to save the result attributes (seenumpy.savez).- Parameters
- path
Path to file in which the result will be saved.
-
property
sensitivities¶ \(d\) vector of normalised HSIC calculation results. These are the main sensitivity results a user should be concerned with. If
n_bootstrapsis larger than one. This represents the mean over the bootstraps.Sensitivities are defined such that:
\[\sum S_d = 1\]\[0 \leq S_d \leq 1\]- See Also
-
property
sensitivities_std¶ \(d\) vector of the standard deviation of normalised HSIC calculation results across all the bootstraps.
- See Also
-
property
-
class
HSICResultWithReweight(*args, **kwargs)[source]¶ Bases:
scm.glompo.analysis.hsic.HSICResult-
property
dg¶ \(n_{bootstraps} \times m\) matrix of gradients of \(g\) with respect to the \(m\) weights in the data set.
-
property
g¶ Metric of the sensitivity imbalance (\(g\)). \(g \geq 0\) such that \(g = 0\) means all sensitivities are on target. If
n_bootstrapsis larger than one. This represents the mean over the bootstraps.
-
property
g_std¶ Standard deviation of \(g\) across all the bootstraps. Returns
Noneif a reweight calculation was not run.
-
property
n_residuals¶ Returns the number of residuals contributing to the loss function.
-
plot_reweight(path='reweightresult.png', group_by=None, labels=None)[source]¶ Create a graphic of the reweight results.
- Parameters
- path
Optional file location in which to save the image. If not provided the image is not saved and only returned.
- group_by
Sequence of integer indices grouping individual loss function contributions by training set item. For example, all the forces in a single data set ‘Forces’ item.
- labels
Sequence of data set item names.
- Returns
- matplotlib.figure.Figure
Figure instance allowing the user to further tweak and change the plot as desired.
-
save_reweight_summary(path='reweightresult.csv', group_by=None, labels=None)[source]¶ Save a summary of the reweight calculation to a text file.
- Parameters
- group_by
Sequence of integer indices grouping individual loss function contributions by training set item. For example, all the forces in a single data set ‘Forces’ item.
- labels
Sequence of data set item names.
-
suggest_weights(group_by=None, return_stats=False)[source]¶ Suggest new weights based on the reweight calculation results. If
n_bootstrapsis larger than one, uses the meandgvalue over the bootstraps.- Parameters
- group_by
Sequence of integer indices grouping individual loss function contributions by training set item. For example, all the forces in a single data set ‘Forces’ item.
-
property
targets¶ \(d\) length vector of target sensitivity values for each parameter. Returns
Noneif a reweight calculation was not run.
-
property
weights¶ Original \(m\) length vector of weights used in the loss function. Returns
Noneif a reweight calculation was not run.
-
property
Kernels¶
Kernels are the key inputs for the sensitivity calculation.
One kernel is applied to the parameters for which the sensitivity is being tested.
One kernel is applied to the function values the sensitivity is being tested against.
Note
Sensitivities are calculated for one parameter at a time, hence the kernels below are not multi-dimensional since their inputs are always scaler values.
-
class
BaseKernel[source]¶ -
__call__(*args)[source]¶ Returns the kernel value or the derivative depending on the number of arguments.
- See Also
-
-
class
ConjunctiveGaussianKernel(gamma=0.1)[source]¶ Bases:
scm.glompo.analysis.kernels.BaseKernelSigmoid type kernel, similar to the
ThresholdKernel, but continuous. Places the bulk of the distribution on thedatapoints with the lowest values.\[k(x, y) = \exp\left[ - \frac{x^2 + y^2}{2\gamma^2} \right]\]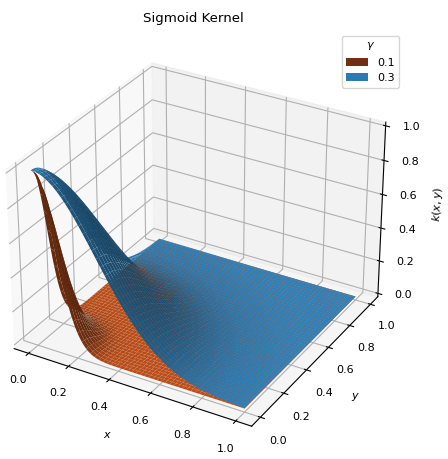
- Parameters
- gamma
Higher values extend the distribution over more points (\(0 < \gamma < 1\)).
-
class
GaussianKernel(sigma=0.3)[source]¶ Bases:
scm.glompo.analysis.kernels.BaseKernelTraditional Gaussian kernel used to measure the similarity between inputs.
\[k(x, y) = \exp\left[ - \frac{(x - y)^2}{2\sigma^2} \right]\]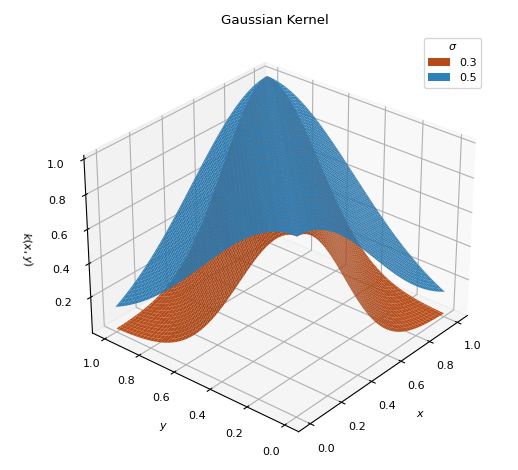
- Parameters
- sigma
Normal distribution bandwidth parameter (\(0 < \sigma < 1\))
-
class
LinearKernel[source]¶ Bases:
scm.glompo.analysis.kernels.PolynomialKernelSpecial instance of
PolynomialKernelof degree 1 and shift 0.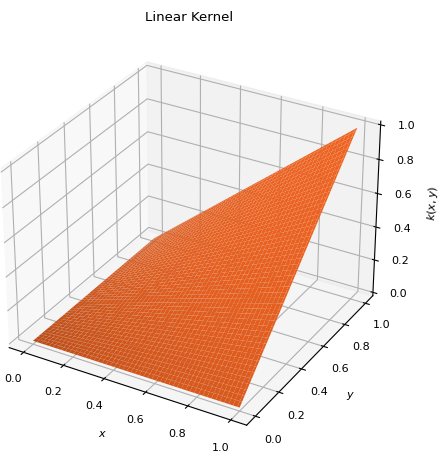 \[k(x, y) = x y\]
\[k(x, y) = x y\]
-
class
PolynomialKernel(order=1, shift=0)[source]¶ Bases:
scm.glompo.analysis.kernels.BaseKernelPolynomial kernel which places the distribution bulk on the highest
datavalues.\[k(x, y) = (x y + c)^d\]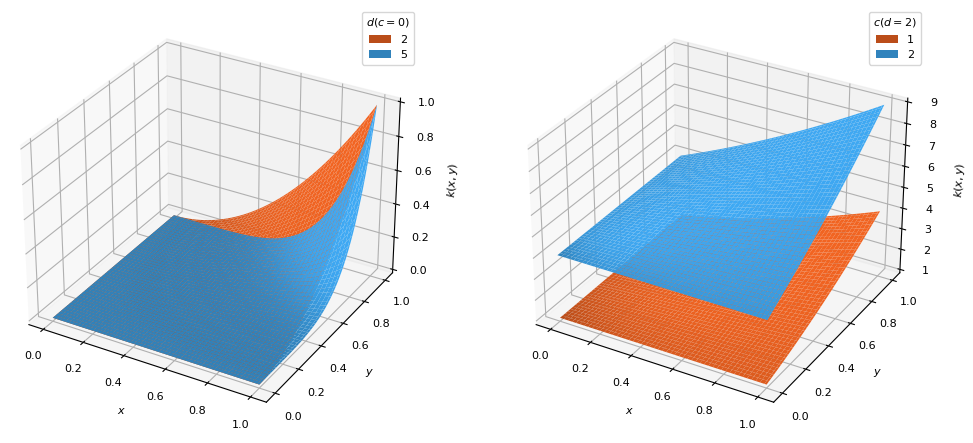
- Parameters
- order
Degree (\(d\)) of the polynomial function (\(d \geq 1\))
- shift
Vertical shift (\(c\)) of the distribution to temper the effects of
order
-
class
ThresholdKernel(alpha)[source]¶ Bases:
scm.glompo.analysis.kernels.BaseKernelSimple linear kernel which first toggles the data to zero or one. Places the distribution over the smallest function values.
\[\begin{split}g(x) = \begin{cases} 1 &\text{ if } x < q_\alpha \\ 0 &\text{ if } x \geq q_\alpha \\ \end{cases}\end{split}\]\[k(x, y) = g(x) g(y)\]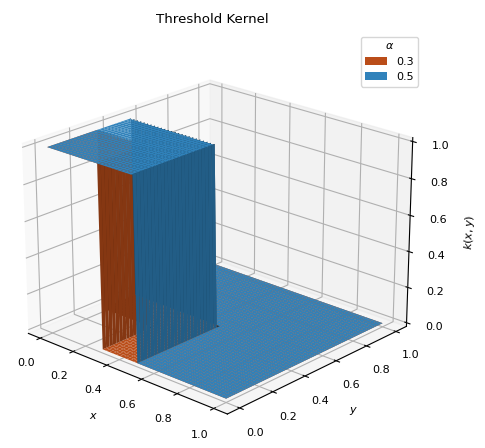
- Parameter
- alpha
Sets all values below the
alphaquantile indatato 1 and all those above to 0 (\(0 < \alpha < 1\)).
- References
Spagnol, A., Riche, R. Le, & Veiga, S. Da. (2019). Global Sensitivity Analysis for Optimization with Variable Selection. SIAM/ASA Journal on Uncertainty Quantification, 7(2), 417–443. https://doi.org/10.1137/18M1167978