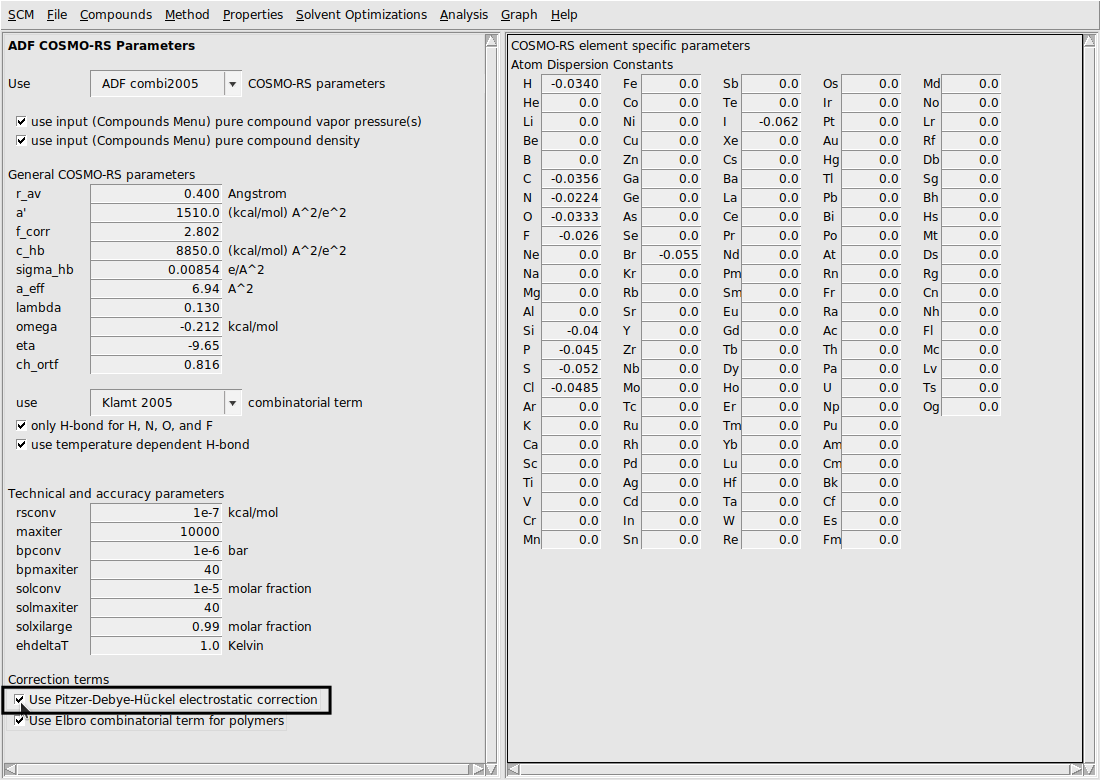
Pitzer-Debye-Hückel electrostatic correction¶
Systems with charged species often exhibit non-ideal behavior – even at low concentrations of ionic components. For this reason, it is often beneficial to include long range electrostatic corrections in the calculation of the activity coefficient. Several models for long range electrostatic energy exist, but one of the most successful and generalizable approaches is the Pitzer-Debye-Hückel (PDH) model [1].
This tutorial sets up a calculation for NaCl in water and compares (molality-based) mean ionic activity coefficients calculated by COSMO-RS to experiment. This tutorial will also perform the same calculation while not considering explicit water solvation (done with the multispecies tools of the ions and additionally while not using the PDH correction.
Turning the PDH correction on/off¶
By default the PDH correction is not used. It can be turned on (or back off) in the Parameters window. If the PDH correction is turned on, it will only have an effect if there are charged species detected in the system. The following steps more exactly describe how to adjust this parameter.
Example: NaCl in Water¶
In this example, we will calculate mean ionic activity coefficients for a very common system: NaCl in water. Though it is possible to calculate the properties of this system with respect to NaCl as a single compound, it is more convenient for calculating mean ionic activity coefficients if we consider Na+ and Cl- as separate compounds. We will perform the calculations using the following 3 approaches:
Using only the bare (unhydrated) ions and no PDH correction
Using explicitly hydrated ions and no PDH correction
Using explicitly hydrated ions and the PDH correction
The user is invited to try to generate the structures necessary for this tutorial, but it is recommended to download the .coskf files in the link above. Using these .coskf files is more convenient and ensures consistency with the tutorial.
Calculating (molality-based) mean ionic activity coefficients¶
As ionic activity coefficients are more commonly reported as mean ionic activity coefficients, we’ll demonstrate how to perform the conversion here. Given a dissociation
where \(v^+\) and \(v^-\) indicate the stoichiometric coefficients of the dissociation, the formula for mean ionic activity coefficients is the following:
In the case of NaCl, this is simply
For electrolytes, the reference state is often taken to be at infinite dilution in the solvent. Of course, in the limit of infinite dilution, the chemical potential goes to \(-\infty\), but we can use the chemical potential calculated with COSMO-RS (the so-called Ben-Naim chemical potential) as it contains no concentration term. We can then calculate the activity coefficients of each species as follows:
where \(\mu_i\) is the chemical potential of an ion calculated by COSMO-RS and \(\mu_i^*\) is the chemical potential of that ion at infinite dilution. Finally, to compare to experimental data, we’ll have to change to a molality scale for activity. That is done as follows:
where \(\gamma_i^{(m)}\) is the molality-based (mean ionic) activity coefficient and \(\gamma_i^{(x)}\) is the mole fraction based activity coefficient. \((\nu^+ + \nu^-)\) is the count of the number of ions the salt dissociates into (2 for NaCl), \(M_s\) is the molar mass of the solvent (in g/mol), and \(m_i\) is the molality of the salt (in mol/kg):
where \(x_i\) is the mole fraction of an ion.
(1) Using unhydrated ions without the PDH correction¶
We’ll first consider the simple case of dissociated Na+ and Cl- ions in water. To set up this experiment, first generate COSMO Result Files for Na+, Cl-, and Water (or simply use the water .coskf file in the database). Then setup a s1-s2 composition line calculation with the following:
This should result in the following

Template calculations for NaCl
For the point \(s1\_x=0.0\) , we have \(\ln(\gamma)\) values for Na+ and Cl- at infinite dilution (these are -45.86 and -56.34, respectively). Next, we subtract these infinite dilution values from all the other \(\ln(\gamma)\) values to convert the standard state to infinite dilution of the ions. In steps, this is:
We refer to the values after the subtraction as \(\ln(\gamma^*)\) values. Using these values, we can calculate the molarity mean ionic activity coefficient as follows:
Finally, this can be converted to molality based units with the following conversion:
Example calculations for the first few points
For the first point at finite dilution (\(s1\_x=0.1\), s1 has 0.1 molar fraction Na+, \(x_{Na+} = 0.1*s1\_x = 0.01\)), we use the \(\ln(\gamma)\) values for Na+ and Cl- (-48.36 and -58.66), the \(\ln(\gamma)^\infty\) values (-45.86 and -56.34), and the mole fraction of Na+ (0.01). These numbers can be used to calculate the molality based mean ionic activity coefficient as follows:
For \(s1\_x=0.1\), we have
For \(s1\_x=0.2\), we have
Repeating this procedure for the remainder of the points results in the following poor correspondence to experiment [2]:

Note that converting the mole fractions to molalities (mol/kg) can be done simply with
where \(M_s\) is the molar mass of the solvent (in g/mol), and \(x_i\) is the mole fraction of Na+.
(2) Using hydrated ions without the PDH correction¶
One of the most important effects in this system is the solvation of the dissociated ions with water molecules. Water molecules interact strongly with the Na+ and Cl- ions in solution, and in this example we will demonstrate that including explicitly solvated species captures the behavior of the system much better than standard COSMO-RS does.
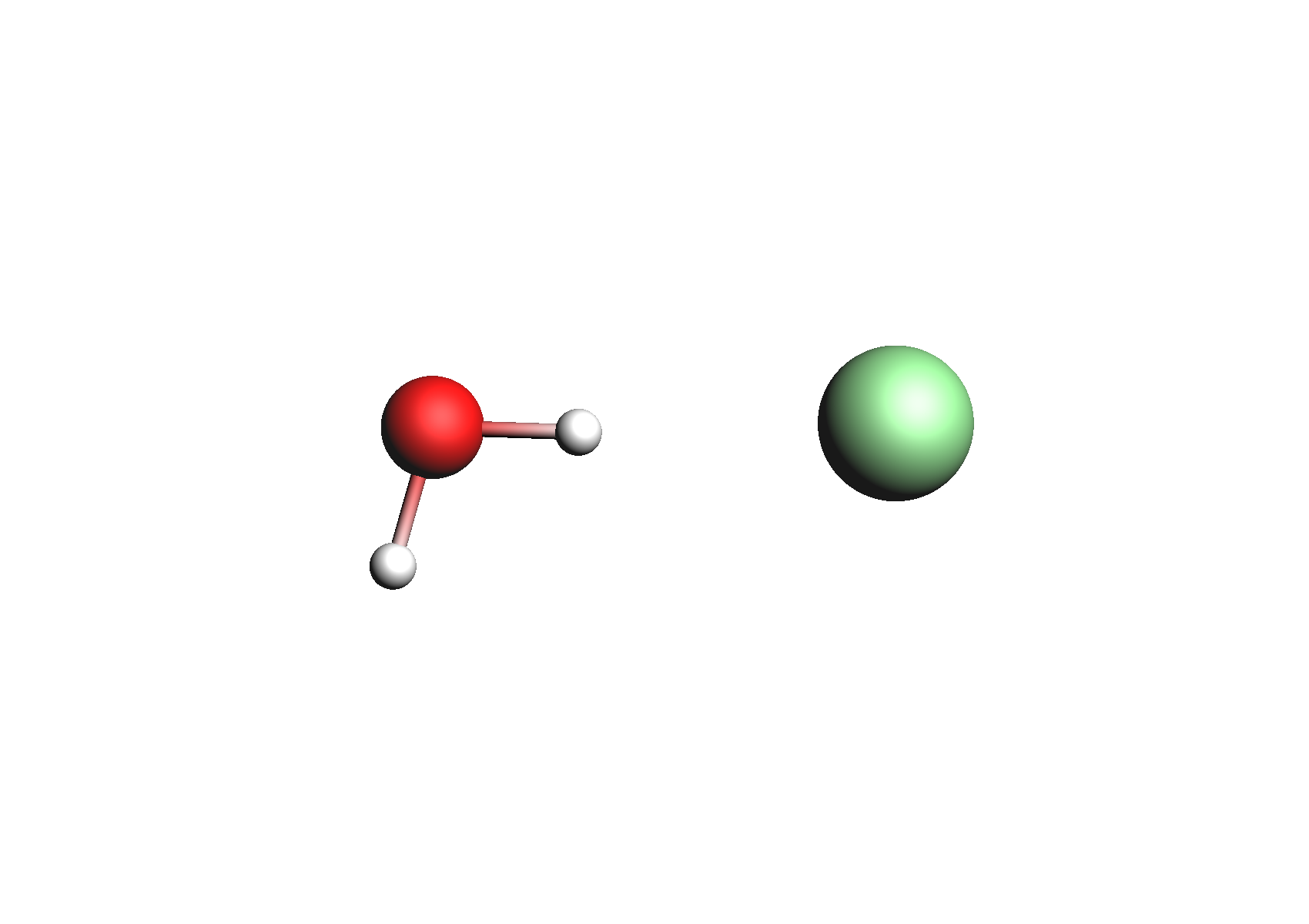
First, we need to do a series of calculations to generate .coskf files for the Na+ and Cl- ions with various degrees of hydration. The geometries of these structures are shown below:





Structures used for the Na+ cation (unassociated and 1,2,3,4 explicit waters)

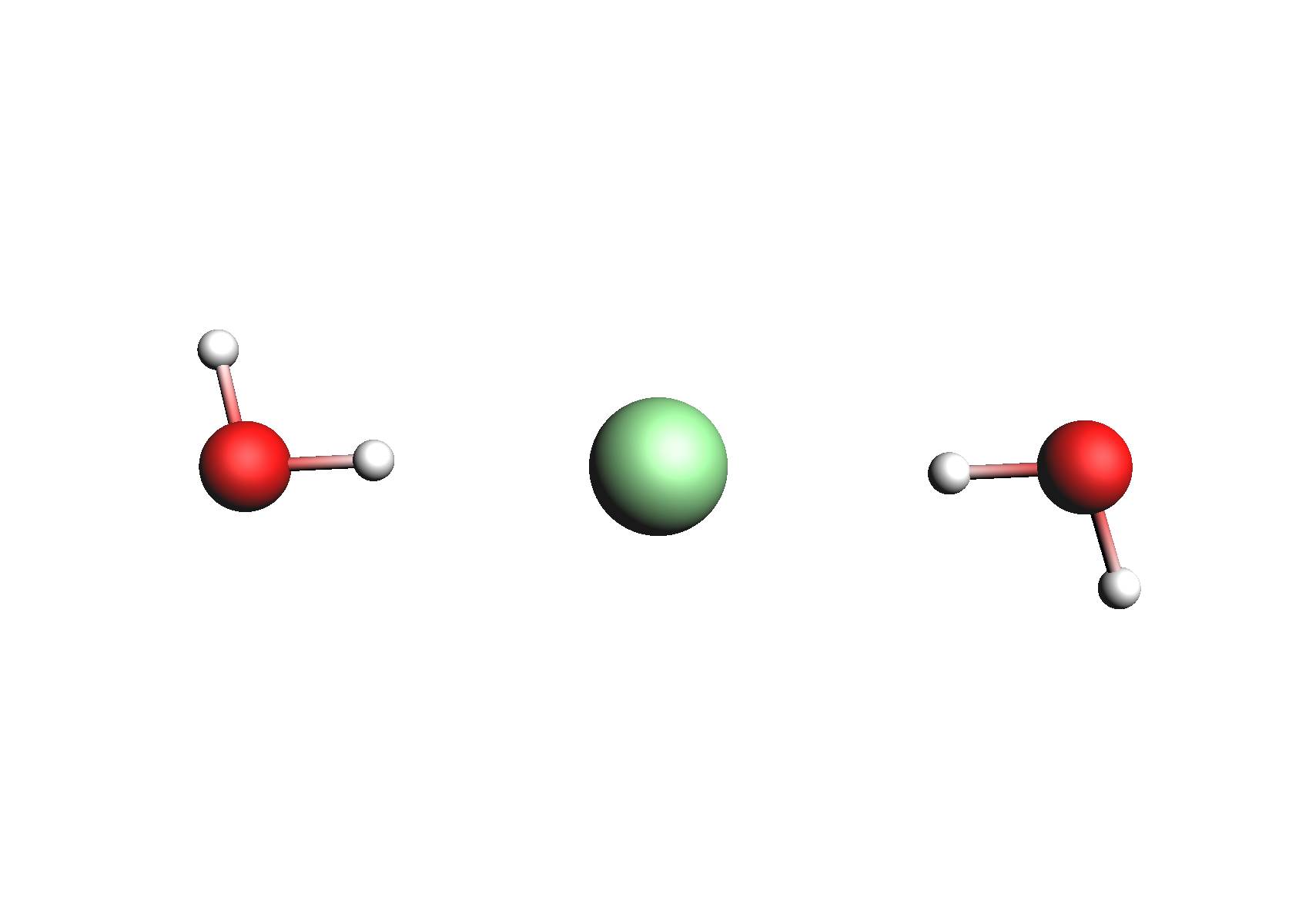
Structures used for the Cl- anion (unassociated and 1,2 explicit waters)
To consider Na+ as a species that can associate, we input the multiform version of Na+:
This should result in the following:

Next, we need to input the multiform Cl-:
Finally, we run calculations with these compounds. We’ll again do a solvent composition line calculation:
This should result in the following:
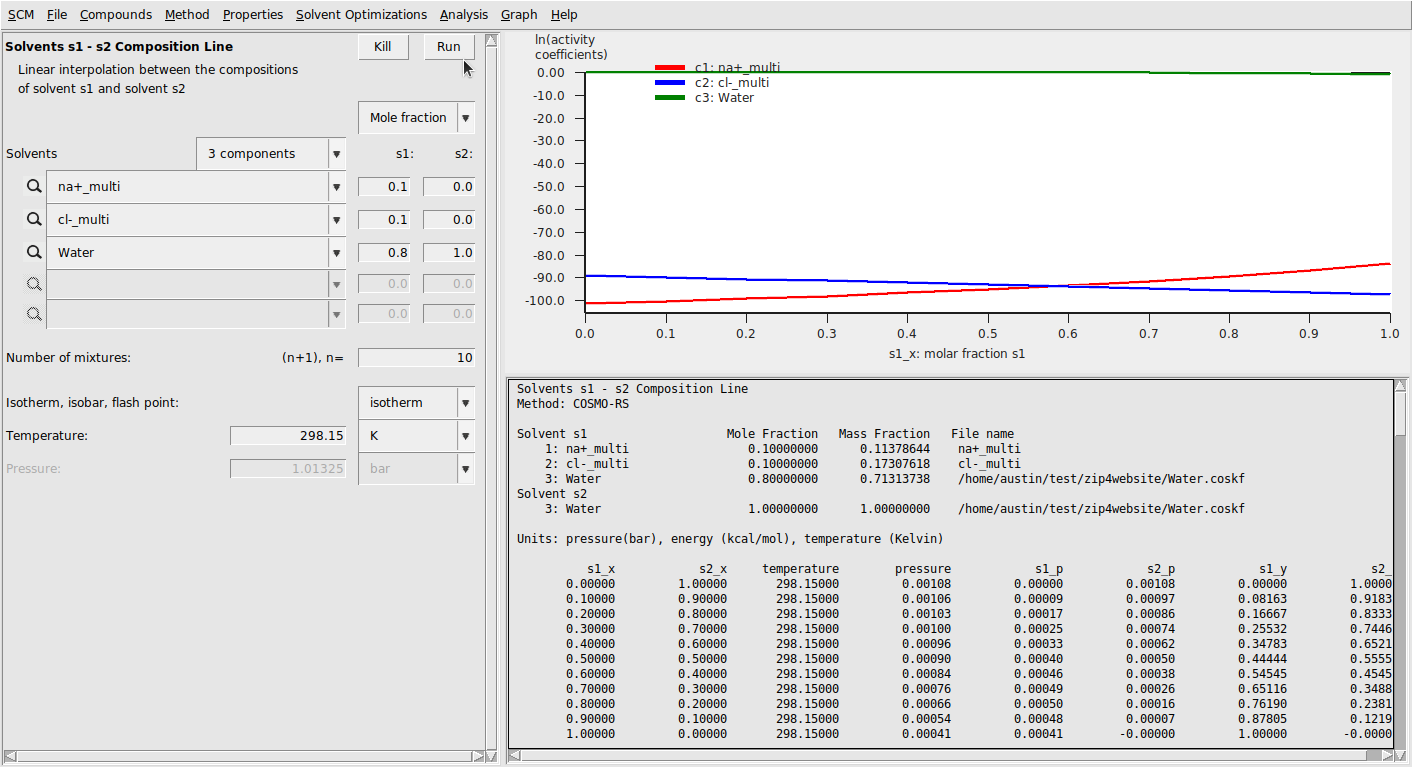
Mean ionic activity coefficients can be calculated with the exact same procedure as above. This results in a much better correspondence to experiment [2], though the results seem to show the wrong trend.

(3) Using hydrated ions with the PDH correction¶
Finally, we use hydrated ions in combination with the PDH correction.
Note
All calculations using the PDH corrections will require densities and dielectric constants for all compounds in the system.
Setting the density and dielectric constant can be done from the Compounds → List of added compounds menu. This input for Na+ looks like the following:

Now, use the following procedure to set up the calculation:
Again following the procedure above to calculate molality-based mean ionic activity coefficients, we obtain a good correspondence to experiment [2], at least for small to moderate ion concentrations.

Additional comments¶
This tutorial provided a basic example of how to add the PDH electrostatic correction for systems with charged species. Attempting to keep the tutorial instructive and easy-to-follow, many considerations for modeling ions with COSMO-RS were left out. Some of these considerations may have improved the quantitative accuracy of this example. A few additional suggestions for modeling small ion/water systems are given below:
The sigma profiles of small ions are very sensitive to the atomic radii. Altering the default radii of small ions (cations in particular) may lead to better results. See [3] for an example of using non-default values for the radii.
Adding correct values for the
HcorrandScorrparameters may also lead to better results. Without these parameters, the fully hydrated species (Na+ with 4 waters and Cl- with 2) are preferred across the whole concentration range. Using these parameters may lead to a more noticeable change in hydration number as a function of ion concentration
References