NEGF: Create a Molecular Junction¶
See also
In this tutorial we will use the NEGF geometry building tools to create a Au-(4,4’-bipyridine)-Au molecular junction:

Instructions¶
Import the leads and fill the central region with 4 layers of lead material:
here to download the lead file Au3x3_lead.xyzWe now carve two tips out of the central gold wire:
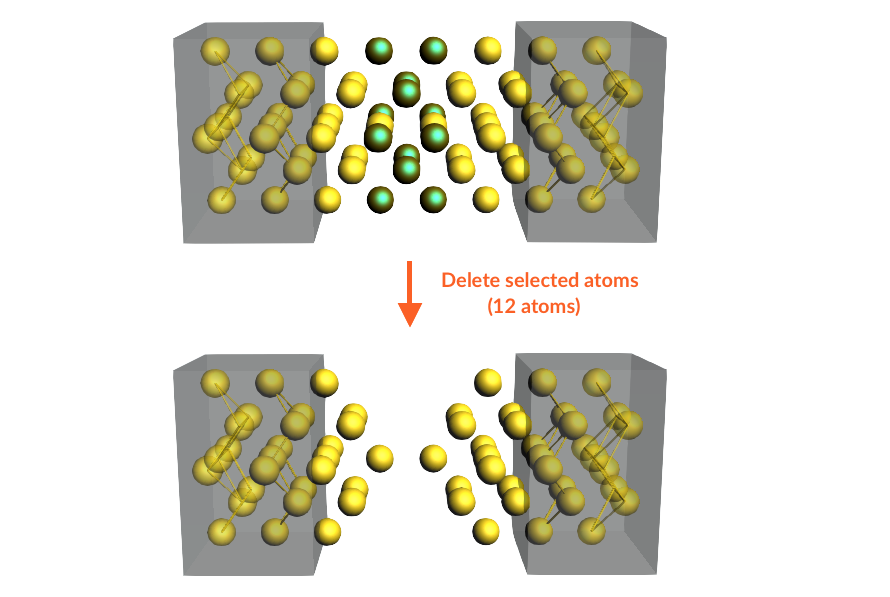
We need to make space in the central region for the 4,4’-bipyridine molecule; to this aim we define a left tip and a right tip:

Your system should now look like this:
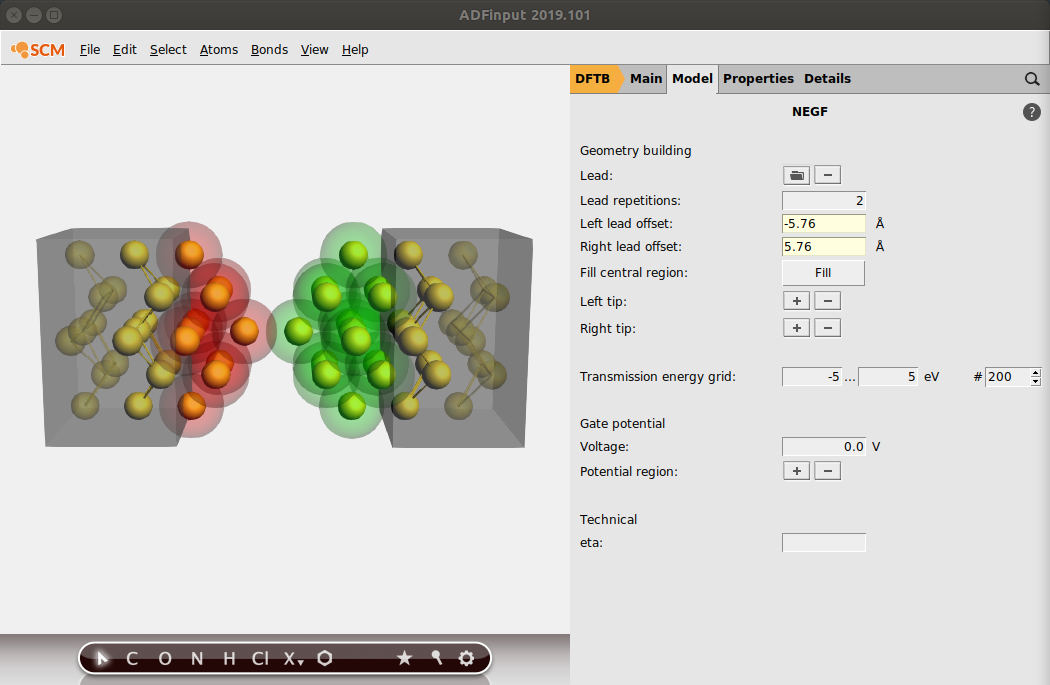
Tip
You can remove atoms from a tip by selecting them and clicking on - next to Left/Right Tip in the NEGF panel
The tips are now anchored to their respective leads. If we move the two leads via the Left/Right lead offset, the tips will follow them.
Make space for 4,4’-bipyridine molecule:
and import it:
here to download the .xyz file 4_4_bipyridine.xyzYour system should now look like this:

Tip
It is good practice to test the convergence of the results with respect to the number of lead repetitions and an amount of buffer lead material in the central region
We are now ready to run the calculation:
Gate potential¶
To include a gate potential for the 4,4’-bipyridine molecule:

We will now run the job and visualize the results:
To better see the effect of the bias potential on the transmission function, we can add the transmission functions at zero gate we computed earlier:



