M3GNet: Universal ML Potential¶
Note: This tutorial requires AMS2023 or later.
This tutorial will teach you how to:
Download and install the M3GNet-UP-2022 machine learning (ML) potential 1 for the entire periodic table, especially suitable for organic and inorganic crystal materials. M3GNet-UP-2022 was fitted to PBE reference data.
Use the D3 Dispersion Engine Addon to add Grimme’s D3 dispersion to the energy, forces, and stress tensors.
Calculate the cohesive energy of the molecular naphthalene crystal.
Note
M3GNet can also be applied to inorganic crystals like metals and metal oxides.
Install M3GNet¶
First, make sure to install the M3GNet package inside AMS.

M3GNet runs much faster on a GPU than a CPU. If you have a CUDA-enabled GPU on your machine:
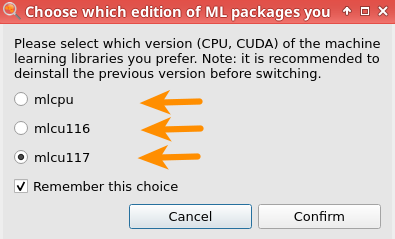
Then, download and install M3GNet:

Cohesive energy¶
To calculate the cohesive energy of solid naphthalene, you need to calculate the energy of
a single naphthalene molecule in the gasphase
the naphthalene crystal
The naphthalene crystal unit cell contains two naphthalene molecules, so ultimately we want to calculate the reaction energy
Gasphase naphthalene¶
naphthalene_gas.xyz
To add D3(BJ) dispersion corrections:

If you have a CUDA-enabled GPU, then you can go to the Details → Technical panel and set Device to cuda:0. However, for a small system like naphthalene, running on a GPU does not provide significant speedup (in practice, the D3 dispersion calculation is about as computationally expensive as the M3GNet calculation).

Then save and run the job:
naphthalene_gas.amsCrystalline naphthalene¶
Set up crystalline naphthalene exactly as the gasphase calculation, with two exceptions:
naphthalene_crystal.xyz instead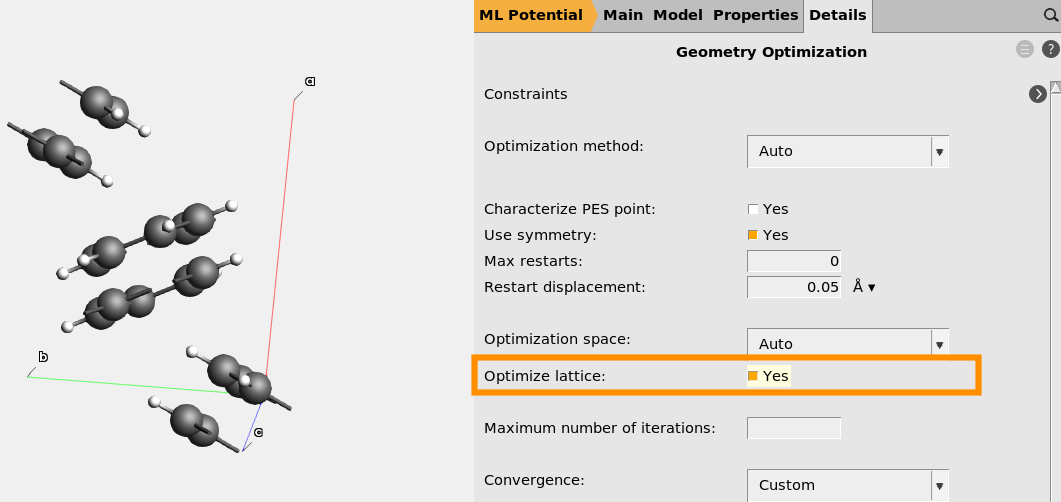
Some of the molecules cross the periodic boundaries, so that the atoms in the same molecule are shown on different sides of the unit cell. This can be confusing. AMSinput offers two ways to make the visualization easier:
This shows some periodic replicas of atoms that are “just outside” the unit cell, but does not change any atomic positions.

Alternatively, you can move the atoms so that all molecules are shown intact (thus placing some atoms outside the unit cell):

It is up to you if you prefer to have the molecules split across the unit cell or not. It makes no difference to the calculation.
Now save with a new name and run the job:
naphthalene_crystal.amsLattice optimization results¶
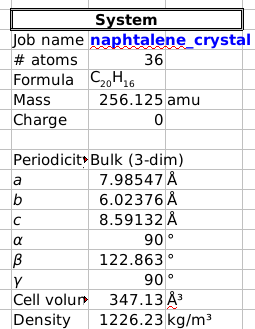
The easiest way to get the optimized lattice parameters is to generate a spreadsheet report:
The optimized cell lengths and angles are shown on the left-hand side.
You can also view the optimized structure in AMSmovie.
Calculate cohesive energy¶
Collect the energies from the gasphase and crystal calculations, either from a spreadsheet as above or by scrolling down towards the end of the .out file (SCM → Output).
You should get Egas = -4.41213477 Ha and Ecrystal = -8.89238031 Ha, giving Ecoh = 0.03406 Ha = 89 kJ/mol.
This compares well to PBE-TS numbers (TS is a different type of dispersion correction) reported by Al-Saidi et al 2:
property |
PBE-TS 2 |
M3GNet-D3(BJ) |
Ecoh (kJ/mol) |
95 |
89 |
a (Å) |
8.01 |
7.99 |
b (Å) |
5.90 |
6.02 |
c (Å) |
8.62 |
8.59 |
Tip
M3GNet is also suitable for inorganic crystals like metals and metal oxides.
M3GNet in Python with PLAMS¶
See the M3GNet example in the PLAMS documentation.
References¶
- 1
C. Chen, S. P. Ong. Nature Computational Science 2, 718–728 (2022). arXiv.2202.02450.
- 2(1,2)
W.A. Al-Saidi, V.K. Voora, K.D. Joradan. DOI: 10.1021/ct200618b

