Adsorption Site Study on Pt(111)¶
This tutorial shows how to use the PES exploration tools of the AMS driver to examine oxygen adsorption sites on a Pt(111) surface using the ReaxFF engine.
Structure Setup¶
We will be using the following structure as basis for the subsequent calculations.
O 5.54003937 1.60484089 8.92740946
Pt 8.31521714 4.80239136 7.78926430
Pt 5.54298786 0.00160612 7.79128800
Pt 6.92910250 2.40199874 7.79027615
Pt 11.08707571 4.80244586 7.78854885
Pt 8.31484643 0.00166062 7.79057254
Pt 9.70096096 2.40178486 7.79068672
Pt 5.54335868 4.80260524 7.78885374
Pt 2.77112940 0.00182001 7.79087743
Pt 4.15724404 2.40221263 7.78986559
Pt 9.70154331 5.60130339 5.52585419
Pt 6.92931403 0.80051815 5.52787789
Pt 8.31542867 3.20091077 5.52686604
Pt 4.15782639 5.60173116 5.52503306
Pt 1.38559711 0.80094593 5.52705675
Pt 2.77171175 3.20133854 5.52604491
Pt 6.92968485 5.60151727 5.52544363
Pt 4.15745557 0.80073204 5.52746732
Pt 5.54357021 3.20112466 5.52645547
Pt 6.92989639 4.00003668 3.26303536
Pt 8.31601103 6.40042930 3.26202352
Pt 5.54378174 1.59964406 3.26404721
Pt 9.70175484 3.99982280 3.26344593
Pt 11.08786948 6.40021541 3.26243408
Pt 8.31564020 1.59943018 3.26445778
Pt 4.15803792 4.00025057 3.26262480
Pt 5.54415256 6.40064318 3.26161295
Pt 2.77192328 1.59985795 3.26363664
Pt 1.38639089 2.39871548 1.00094199
Pt 5.54436409 4.79916260 0.99921469
Pt 6.93024583 7.19969029 1.00043291
Pt 4.15824945 2.39876998 1.00022653
Pt 8.31622256 4.79894871 0.99962525
Pt 9.70210429 7.19947641 1.00084348
Pt 6.93010792 2.39855609 1.00063710
Pt 2.77250564 4.79937648 0.99880412
Pt 4.15838737 7.19990418 1.00002234
VEC1 8.31557575 0.00000000 0.00000000
VEC2 4.15778787 7.20149984 0.00000000
Hint
To build a custom crystal surface slab model, check out the Tutorial on Building Crystals and Slabs
PES Exploration¶
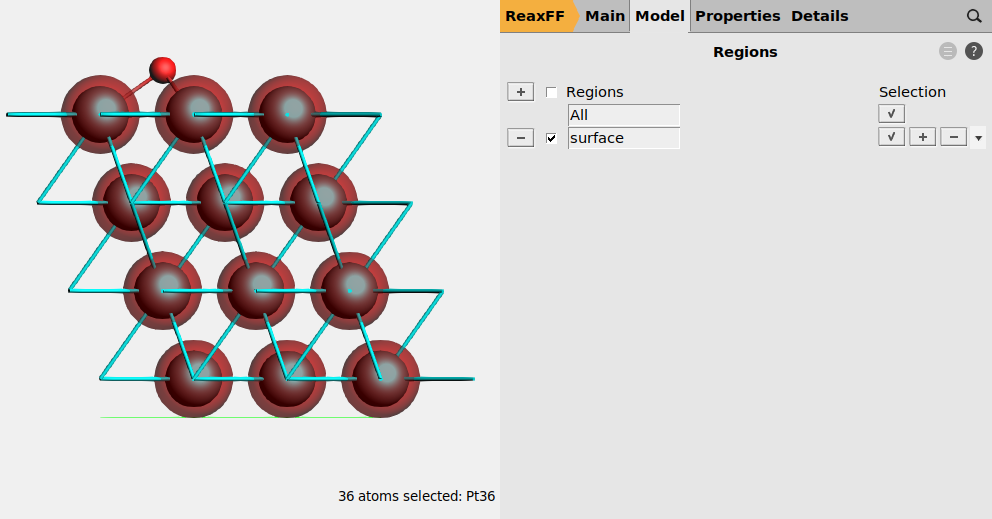
With the Pt(111) model in place, we can proceed to set up the rest of the calculation parameters.
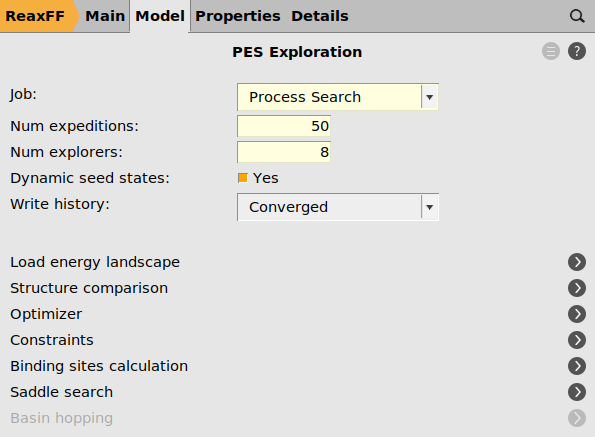
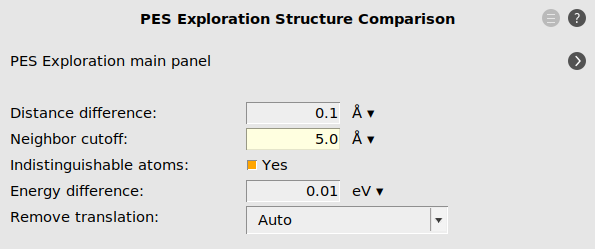
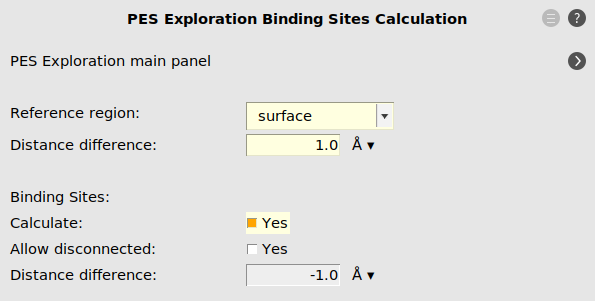
As we are only interested in finding binding sites, i.e. local minima accessible by surface hopping reactions, we have to limit the barrier heights allowed during the process search

Computation & Results¶
We are now ready to start the calculation
The calculation should take a couple of minutes, during which we can monitor the discovery of new minima and transition states either from  → logfile
→ logfile
The newly discovered states can also be inspected in  → Movie
→ Movie

where we can see the nicely resolved energy differences between the two different hollow sites on an fcc (111) surface. Note, that in the case of oxygen adsorption, bridge sites act as transition states for the hopping between different hollow sites, while top adsorption sites are too high in energy to be reached in this process search run.
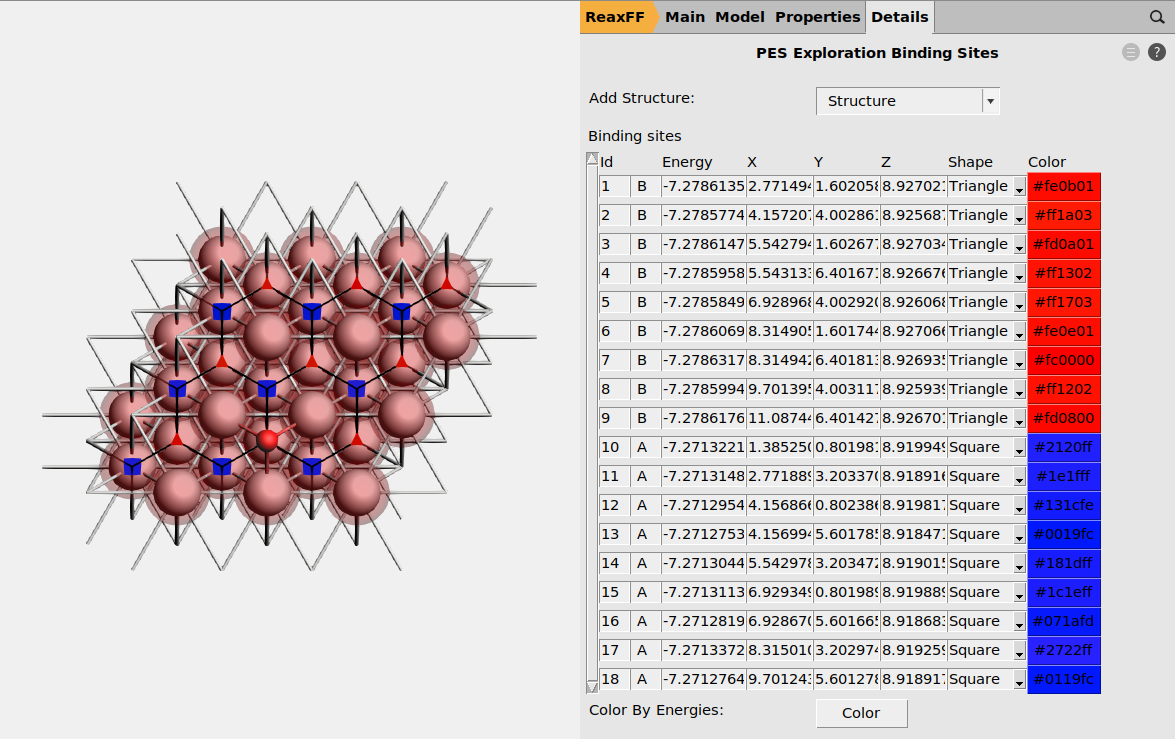
After the calculation is concluded, we return to  → Input and click Yes in the prompt to create a new job and load binding sites.
→ Input and click Yes in the prompt to create a new job and load binding sites.
This displays the identified adsorption sites, which can be individually selected from Details → PES Exploration Binding Sites