PEDA + NOCV - decomposing the orbital relaxation term¶
This tutorial will teach you how to:
- perform a periodic energy decomposition analysis (PEDA) combined with the natural orbitals for chemical valency (NOCV) method for extended systems with the BAND-GUI (PEDA-NOCV)
- where to look for the results
- how to visualize NOCV orbitals and NOCV deformation densities
Step 1: Setting up the System and the Calculation¶
Do Steps 1 to 3c of the general PEDA Tutorial first.
Step 1a: Preparations for the PEDA-NOCV calculation¶
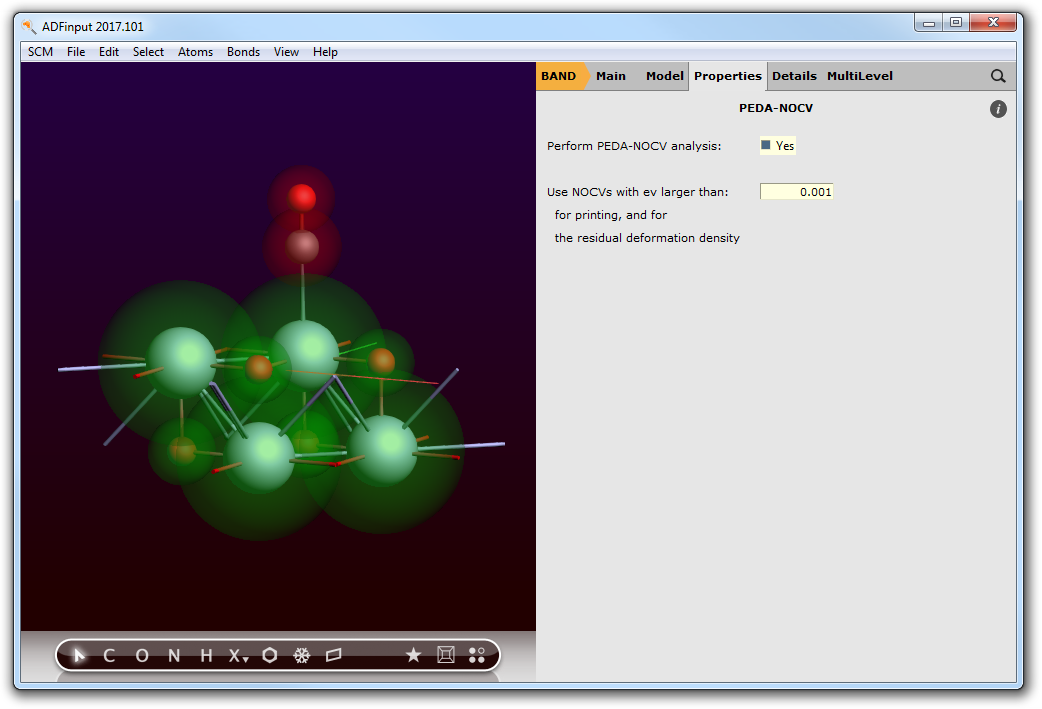
Now the PEDA-NOCV has to be switched on. Go to the PEDA-NOCV menu,
- Panel bar Properties → PEDA-NOCV
and activate the calculation of everything connected to the PEDA-NOCV. You may want to define a NOCV eigenvalue threshold (default: 0.001) which handles the amount of output.
Then you have to keep in mind, that the PEDA-NOCV is implemented for \(\Gamma\)-only case. So, go to the ‘Integration K-Space’ menu,
- Panel bar Details → Integration K-Space
and change the quality of the K-space to be ‘Gamma only’.

This is possible by defining a (1x1)-grid, too.
Step 1b: Save and run the calculation¶
Now you can save and run the calculation.
- File → Save, give it a name and press Save.File → Run
Step 2: Checking the results¶
After the calculations of the fragments and the PEDA finished you can look for the PEDA results. Therefore, open the “Output” using the SCM dropdown menu.
- SCM → Output
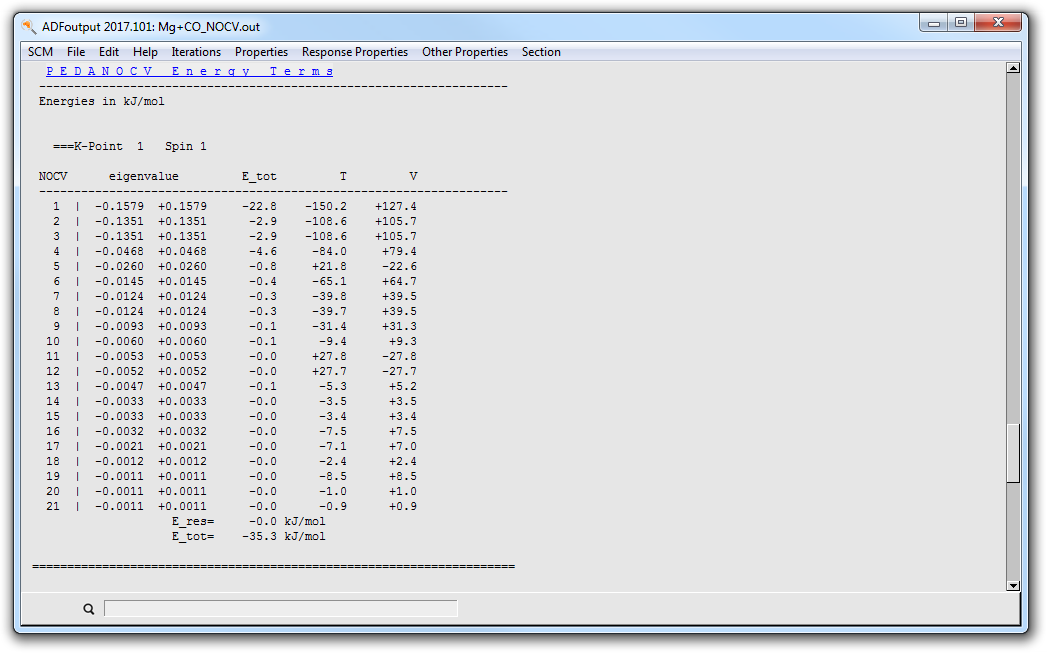
You can jump to the ‘PEDA-NOCV Energy Terms’ via the corresponding button in the ‘Properties’ dropdown menu.
- Properties → PEDA-NOCV Energy Terms
Reference results:
Step 3: Plotting NOCV orbitals and deformation densities¶
You can visualize the charge NOCV deformation densities which describe the charge flow between the fragments. Therefore, open the “View” using the SCM dropdown menu.
- SCM → ViewFields → Grid → Medium
Depending on your preferences (w.r.t. showing atoms of neighboring cells) you will end up with the following representation of ADFview.

You can now add an isosurface (Double(+/-)) via the dropdown menu ‘Add’, or via ‘Ctrl+D’.
- Add → Isosurface (Double(+/-)

Step 3a: Plotting NOCV deformation densities¶
To get an overview of the NOCV deformation densities click on ‘Select Field ...’ and then on ‘NOCV deformation densities’. The following window should appear.

For example you can now visualize the first NOCV deformation density. (Be aware of the small isovalue of 0.000025 in the following picture!)
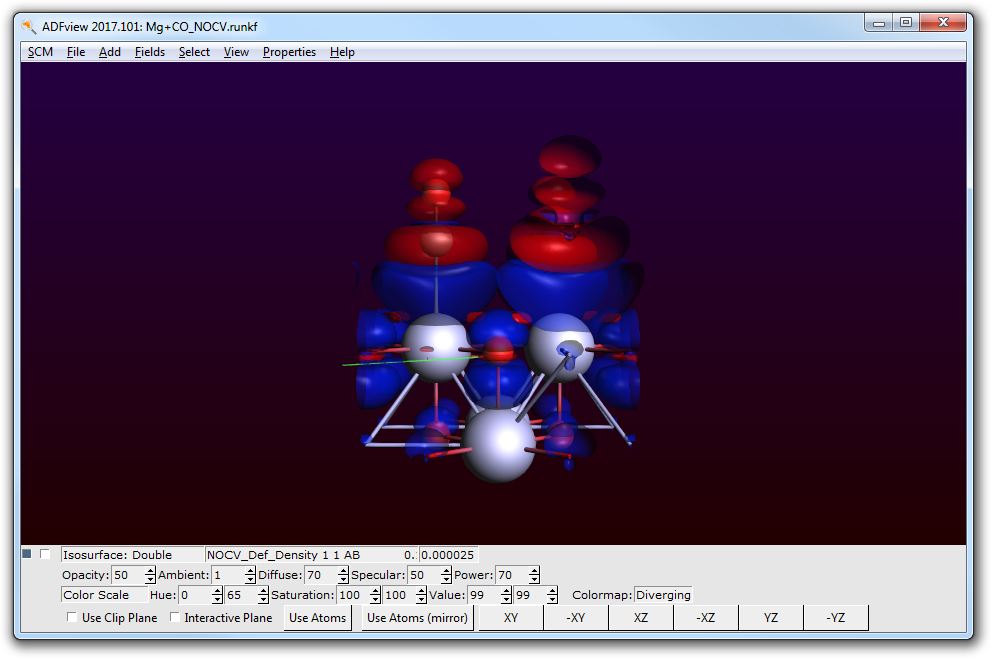
This deformation density visualizes the charge flow from red to blue lobes. Here, the charge transfer from the CO to the MgO surface is shown.
According to the information in the ‘Select NOCV Deformation Density’ window the first NOCV deformation density is a combination of the NOCV orbitals 3 (the small number denotes the donor/occupied NOCV) and 150 (the large number denotes the acceptor/unoccupied NOCV).

Step 3b: Plotting NOCV orbitals¶
These NOCV orbitals can visualized by changing the field to ‘NOCV Orbitals’. The following window should appear.

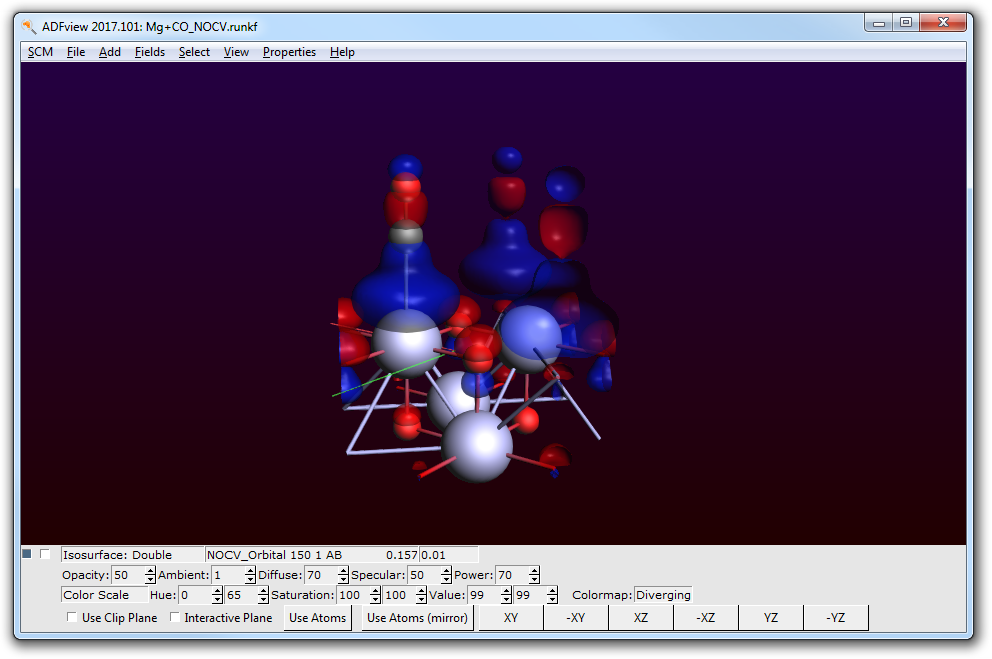
By selecting the 1st and 150th NOCV orbital these are calculated and can then be visualized one at a time.
NOCV orbital 1 does show predominantly a lone-pair localized at the CO fragment.

NOCV orbital 150 does show predominantly a lobe connecting the CO fragment and a Mg atom of the surface.
Remark: The calculated fields for the NOCV properties does represent not only the central unit cell but partially the neighboring cells. So, it can be tough to only show what is needed and not confuse additional lobes of the neighboring cells with something in the unit cell! You can hide parts of the field using the Clip Plane in BANDGUI.
- Isosurface: Double → DetailsCheck Use Clip Plane
To optimize the resulting plot one can visualize (temporarily) and modify the position of the Clip Plane by checking Interactive Plane.
- Check Interactive Plane
